Get the best of both worlds with Fiji's Jython interpreter
Among the plugin smorgasbord of Fiji is the Bio-Formats importer, which can open any proprietary microscopy file under the sun. (And there’s a lot of them!) Below I will use Jython to open some .lifs, do some processing, and output some .pngs that I can process further using Python/NumPy/scikit-image. (A .lif is a Leica Image File, because there were not enough image file formats before Leica came along.)
The first thing to note is that Jython is not Python, and it is certainly not Python 2.7. In fact, the Fiji Jython interpreter implements Python 2.5, which means no argparse. Not to worry though, as argparse is implemented in a single, pure Python file distributed under the Python license. So:
Tip #1: copy argparse.py into your project.
This way you’ll have access the state of the art in command line argument processing from within the Jython interpreter.
To get Fiji to run your code, you simply feed it your source file on the command line. So, let’s try it out with a simple example, echo.py:
import argparse
if name == 'main':
parser = argparse.ArgumentParser(description=
"Parrot back your arguments.")
parser.add_argument('args', nargs="*", help="The input arguments.")
args = parser.parse_args()
for arg in args.args:
print(arg)
$ fiji echo.py hello world
hello
world
But sadly, Fiji captures any -h calls, which defeats the purpose of using argparse in the first place!
$ fiji echo.py -h
Usage: /Applications/Fiji.app/Contents/MacOS/fiji-macosx [<Java options>.. --] [<ImageJ options>..] [<files>..]
Java options are passed to the Java Runtime, ImageJ
options to ImageJ (or Jython, JRuby, ...).
In addition, the following options are supported by ImageJ:
General options:
--help, -h
show this help
--dry-run
show the command line, but do not run anything
--debug
verbose output
(… and so on, the output is quite huge.)
(Note also that I aliased the Fiji binary, that long path under /Applications, to a simple fiji command; I recommend you do the same.)
However, we can work around this by calling help using Python as the interpreter, and only using Fiji to actually run the file:
$ python echo.py -h
usage: echo.py [-h] [args [args ...]]
Parrot back your arguments.
positional arguments:
args The input arguments.
optional arguments:
-h, --help show this help message and exitThat’s more like it! Now we can start to build something a bit more interesting, for example, something that converts arbitrary image files to png:
import argparse
from ij import IJ # the IJ class has utility methods for many common tasks.
def convert_file(fn):
"""Convert the input file to png format.
Parameters
----------
fn : string
The filename of the image to be converted.
"""
imp = IJ.openImage(fn)
# imp is the common name for an ImagePlus object,
# ImageJ's base image class
fnout = fn.rsplit('.', 1)[0] + '.png'
IJ.saveAs(imp, 'png', fnout)
if name == 'main':
parser = argparse.ArgumentParser(description="Convert TIFF to PNG.")
parser.add_argument('images', nargs='+', help="Input images.")
args = parser.parse_args()
for fn in args.images:
convert_file(fn)
Boom, we’re done. But wait, we actually broke the Python interpreter compatibility, since ij is not a Python library!
$ python convert2png.py -h
Traceback (most recent call last):
File "convert.py", line 2, in <module>
from ij import IJ # the IJ class has utility methods for many common tasks.
ImportError: No module named ij
Which brings us to:
Tip #2: only import Java API functions within the functions that use them.
By moving the from ij import IJ statement into the convert function, we maintain compatibility with Python, and can continue to use argparse’s helpful documentation strings.
Next, we want to use the Bio-Formats importer, which is class BF in loci.plugins. Figuring out the class hierarchy for arbitrary plugins is tricky, but you can find it here for core ImageJ (using lovely 1990s-style frames) and here for Bio-Formats, and Curtis Rueden has made this list for other common plugins.
When you try to open a file with Bio-Formats importer using the Fiji GUI, you get the following dialog:
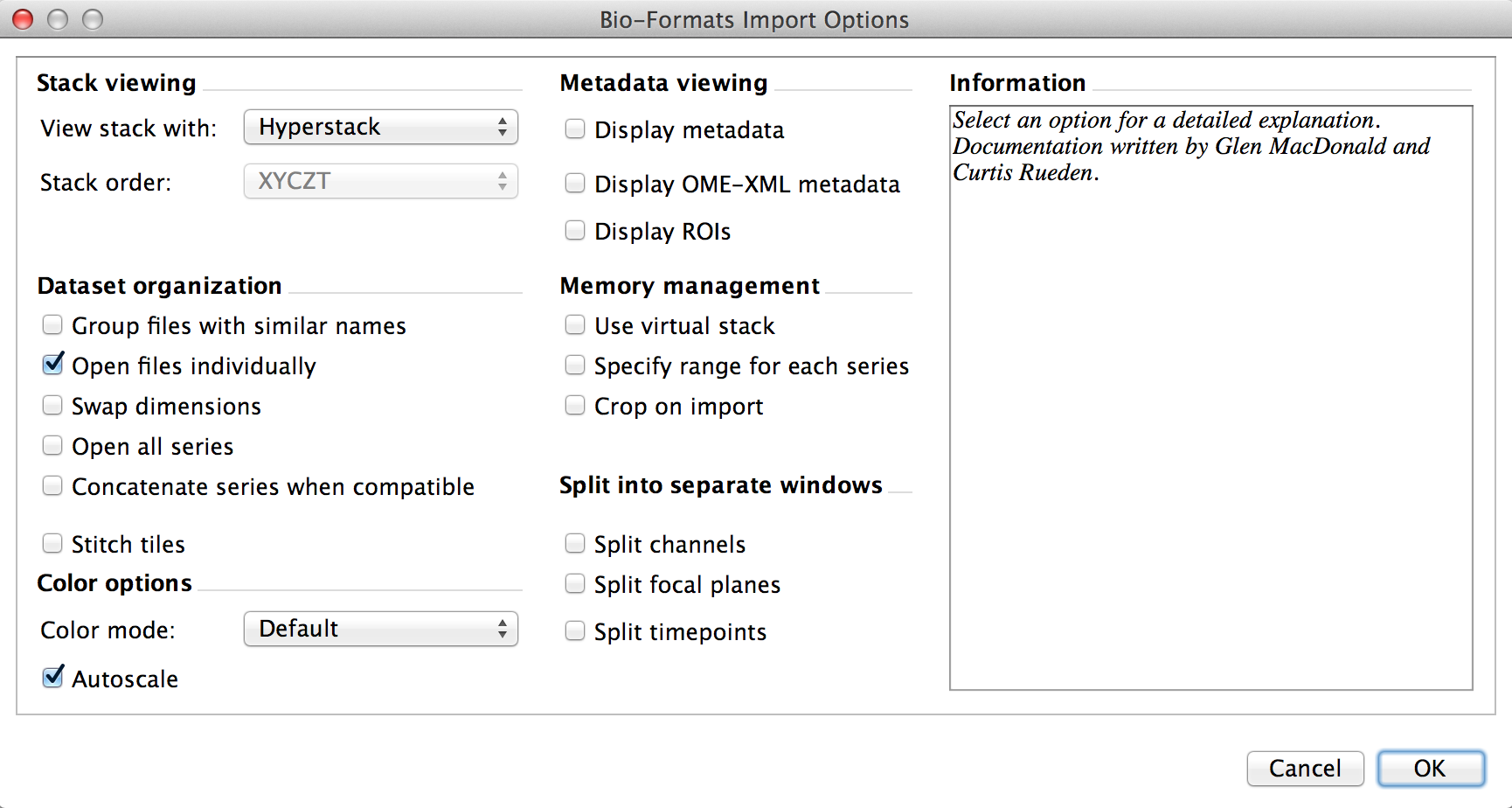
That’s a lot of options, and we actually want to set some of them. If you look at the BF.openImagePlus documentation, you can see that this is done through an ImporterOptions class located in loci.plugins.in. You’ll notice that “in” is a reserved word in Python, so from loci.plugins.in import ImporterOptions is not a valid Python statement. Yay! My workaround:
Tip #3: move your Fiji imports to an external file.
So I have a jython_imports.py file with just:
from ij import IJ
from loci.plugins import BF
from loci.plugins.in import ImporterOptions
Then, inside the convert_files() function, we just do:
from jython_imports import IJ, BF, ImporterOptions
This way, the main file remains Python-compatible until the convert() function is actually called, regardless of whatever funky and unpythonic stuff is happening in jython_imports.py.
Onto the options. If you untick “Open files individually”, it will open up all matching files in a directory, regardless of your input filename! Not good. So now we play a pattern-matching game in which we match the option description in the above dialog with the ImporterOptions API calls. In this case, we setUngroupFiles(True). To specify a filename, we setId(filename). Additionally, because we want all of the images in the .lif file, we setOpenAllSeries(True).
Next, each image in the series is 3D and has three channels, but we are only interested in a summed z-projection of the first channel. There’s a set of ImporterOptions methods tantalizingly named setCBegin, setCEnd, and setCStep, but this is where I found the documentation sorely lacking. The functions take (int s, int value) as arguments, but what’s s??? Are the limits closed or open? Code review is a wonderful thing, and this would not have passed it. To figure things out:
Tip #4: use Fiji’s interactive Jython interpreter to figure things out quickly.
You can find the Jython interpreter under Plugins/Scripting/Jython Interpreter. It’s no IPython, but it is extremely helpful to answer the questions I had above. My hypothesis was that s was the series, and that the intervals would be closed. So:
>>> from loci.plugins import BF >>> from loci.plugins.in import ImporterOptions >>> opts = ImporterOptions() >>> opts.setId("myFile.lif") >>> opts.setOpenAllSeries(True) >>> opts.setUngroupFiles(True) >>> imps = BF.openImagePlus(opts)
Now we can play around, with one slight annoyance: the interpreter won’t print the output of your last statement, so you have to specify it:
>>> len(imps) >>> print(len(imps)) 18
Which is what I expected, as there are 18 series in my .lif file. The image shape is given by the getDimensions() method of the ImagePlus class:
>>> print(imps[0].getDimensions()) array('i', [1024, 1024, 3, 31, 1]) >>> print(imps[1].getDimensions()) array('i', [1024, 1024, 3, 34, 1])</code></pre> That’s (x, y, channels, z, time).
Now, let’s try the same thing with setCEnd, assuming closed interval:
>>> opts.setCEnd(0, 0) ## only read channels up to 0 for series 0? >>> opts.setCEnd(2, 0) ## only read channels up to 0 for series 2? >>> imps = BF.openImagePlus(opts) >>> print(imps[0].getDimensions()) array('i', [1024, 1024, 1, 31, 1]) >>> print(imps[1].getDimensions()) array('i', [1024, 1024, 3, 34, 1]) >>> print(imps[2].getDimensions()) array('i', [1024, 1024, 1, 30, 1])
Nothing there to disprove my hypothesis! So we move on to the final step, which is to z-project the stack by summing the intensity over all z values. This is normally accessed via Image/Stacks/Z Project in the Fiji GUI, and I found the corresponding ij.plugin.ZProjector class by searching for “proj” in the ImageJ documentation. A ZProjector object has a setMethod method that usefully takes an int as an argument, with no explanation in its docstring as to which int translates to which method (sum, average, max, etc.). A little more digging in the source code reveals some class static variables, AVG_METHOD, MAX_METHOD, and so on.
Tip #5: don’t be afraid to look at the source code. It’s one of the main advantages of working in open-source.
So:
>>> from ij.plugin import ZProjector >>> proj = ZProjector() >>> proj.setMethod(ZProjector.SUM_METHOD) >>> proj.setImage(imps[0]) >>> proj.doProjection() >>> impout = proj.getProjection() >>> print(impout.getDimensions()) array('i', [1024, 1024, 1, 1, 1])
The output is actually a float-typed image, which will get rescaled to [0, 255] uint8 on save if we don’t fix it. So, to wrap up, we convert the image to 16 bits (making sure to turn off scaling), use the series title to generate a unique filename, and save as a PNG:
>>> from ij.process import ImageConverter
>>> ImageConverter.setDoScaling(False)
>>> conv = ImageConverter(impout)
>>> conv.convertToGray16()
>>> title = imps[0].getTitle().rsplit(" ", 1)[-1]
>>> IJ.saveAs(impout, 'png', "myFile-" + title + ".png")
You can see the final result of my sleuthing in lif2png.py and jython_imports.py. If you would do something differently, pull requests are always welcome.
Before I sign off, let me recap my tips:
-
copy argparse.py into your project;
-
only import Java API functions within the functions that use them;
-
move your Fiji imports to an external file;
-
use Fiji’s interactive Jython interpreter to figure things out quickly; and
-
don’t be afraid to look at the source code.
And let me add a few final comments: once I started digging into all of Fiji’s plugins, I found documentation of very variable quality, and worse, virtually zero consistency between the interfaces to each plugin. Some work on “the currently active image”, some take an ImagePlus instance as input, and others still a filename or a directory name. Outputs are equally variable. This has been a huge pain when trying to work with these plugins.
But, on the flipside, this is the most complete collection of image processing functions anywhere. Along with the seamless access to all those functions from Jython and other languages, that makes Fiji very worthy of your attention.
Acknowledgements
This post was possible thanks to the help of Albert Cardona, Johannes Schindelin, Wayne Rasband, and Jan Eglinger, who restlessly respond to (it seems) every query on the ImageJ mailing list. Thanks!
References
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, & Cardona A (2012). Fiji: an open-source platform for biological-image analysis. Nature methods, 9 (7), 676-82 PMID: 22743772
Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, Loranger B, Moore J, Neves C, Macdonald D, Tarkowska A, Sticco C, Hill E, Rossner M, Eliceiri KW, & Swedlow JR (2010). Metadata matters: access to image data in the real world. The Journal of cell biology, 189 (5), 777-82 PMID: 20513764
Comments
Comments powered by Disqus